Downtown Walk and Tornado Exibit
Meet at 11:30 in the lobby of Patterson Office Tower.
Bring money for eating lunch downtown.
Email Lina Crocker at lina.crocker@uky.edu if you are attending.
Meet at 11:30 in the lobby of Patterson Office Tower.
Bring money for eating lunch downtown.
Email Lina Crocker at lina.crocker@uky.edu if you are attending.
---------
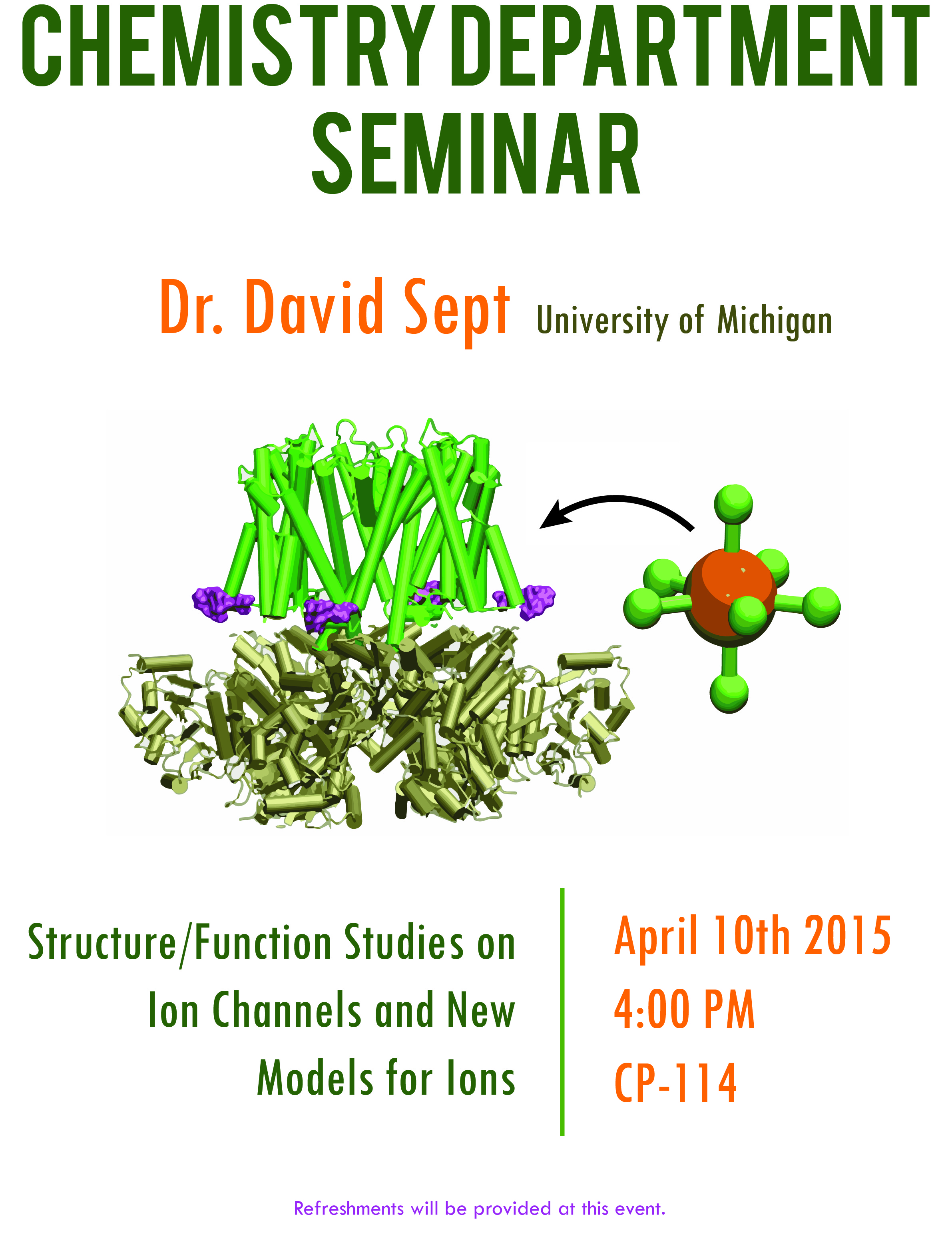
Dr. David Sept will be presenting a seminar titled Structure/Function Studies on Ion Channels and New Models for Ions. Refreshments will be provided at this event.
Abstract: Voltage and Ca2+ activated BK channels modulate neuronal activities. Previous studies found that the Ca2+ binding sites and the activation gate are spatially separated, but exactly how Ca2+ binding couples to gate opening is not clear. We address this question by studying how a mutation in BK channels, which is associated with generalized epilepsy and paroxysmal dyskinesia, enhances Ca2+ sensitivity. This epilepsy mutation is located in a structural domain (the AC region) that is close to a putative Ca2+ binding site, and mutagenesis studies show that the AC region is important in the coupling between Ca2+ binding and gate opening. Through a combination of experimental and computational studies, we find the epilepsy mutation enhances Ca2+ sensitivity by an allosteric mechanism affecting the coupling between Ca2+ binding and gate opening.
Faculty Host: Dr. Pete Kekenes-Huskey
Title: Oscillatory Riemann Hilbert
Abstract: Please see the seminar web page at http://www.ms.uky.edu/~hislop/PDEseminarS2015
In the fall of 2014, I will begin doctoral studies in anthropology at Yale University. This ambition to further advance my education in archaeology was forged while completing my baccalaureate degree at the University of Kentucky.
We are having a meeting on Tuesday, March 30 at 5:15pm to discuss the details of our invited speaker for the semester.
If you have further questions please contact one of the following people.
![]()
George Lytle
Chase Russell
Fouche Smith
Title: Optimality of the Neighbor Joining Algorithm and Faces of the Balanced Minimum Evolution Polytope
Abstract: Balanced minimum evolution (BME) is a statistically consistent distance-based method to reconstruct a phylogenetic tree from an alignment of molecular data. In 2008, Eickmeyer, Huggins, Pachter, and myself developed a notion of the BME polytope, the convex hull of the BME vectors obtained from Pauplin's formula applied to all binary trees. We also showed that the BME can be formulated as a linear programming problem over the BME polytope. The BME is related to the Neighbor Joining (NJ) algorithm, now known to be a greedy optimization of the BME principle. Further, the NJ and BME algorithms have been studied previously to understand when the NJ algorithm returns a BME tree for small numbers of taxa. In this talk we aim to elucidate the structure of the BME polytope and strengthen knowledge of the connection between the BME method and NJ algorithm. We first show that any subtree-prune-regraft move from a binary tree to another binary tree corresponds to an edge of the BME polytope. Moreover, we describe an entire family of faces parametrized by disjoint clades. We show that these clade-faces are smaller-dimensional BME polytopes themselves. Finally, we show that for any order of joining nodes to form a tree, there exists an associated distance matrix (i.e., dissimilarity map) for which the NJ algorithm returns the BME tree. More strongly, we show that the BME cone and every NJ cone associated to a tree T have an intersection of positive measure. We end this talk with the current and future projects on phylogenomics with biologists in University of Kentucky and Eastern Kentucky University. This work is supported by NIH.
----------
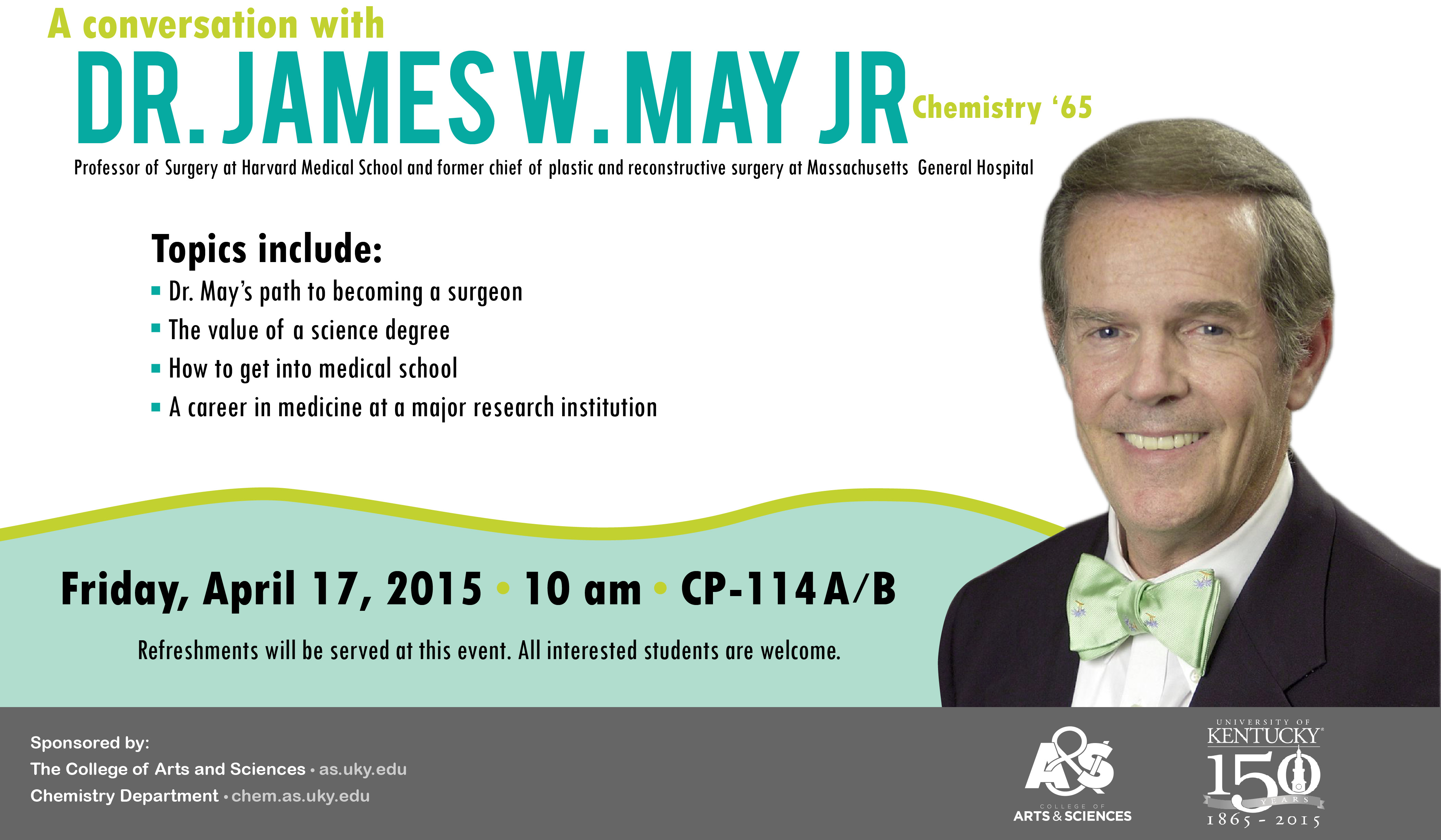
Dr. James W. May Jr (Chemistry '65) will be available to talk with students about several topics, including: his path to becoming a surgeon, the value of a science degree, how to get into medical school, and a career in medicine at a major research institution.
All interested students are welcome. Refreshments will be served at this event.
Co-sponsored by the College of Arts & Sciences and the Department of Chemistry